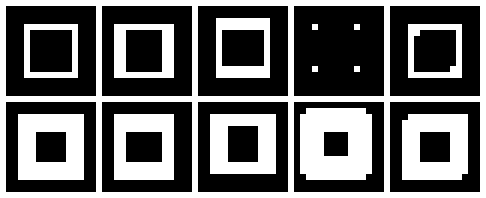
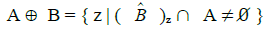Sometimes, we would like to get measurements from images and the process would be easier if the region of interest (ROI) is clearly distinguishable from the rest of the image. In other words, we want our ROI to be well-segmented from its background and it can be achieved by edge detection or by specifying the ROI as a blob. Either way, the images must be first undergo preprocessing in order to make them more suitable for performing the desirable measurements. This activity makes use of blobs to analyze the 'cells' found in the image.
Determining the best estimate of area of the cells
In image processing, blob is considered to be a region of connected pixels. In Scilab, blobs are detected from binary images which exclude them to the background pixels or those having zero values. You can see below the image of white circles which we consider the cells
To process the images easier and faster, it was cropped into 256x256 pixels subimages. The image was cropped using GIMP and we allowed overlapping subimages. This resulted to 12 subimages.
One subimage (upper left part of the whole image) is shown below.
 |
| Subimage 1. |
To segment our regions of interest, we need to binarize the image. We have to consider the histogram of the image in order to determine the enough threshold value to produce an output image that will best isolate our cells and background. I first used the usual imhist() function and obtain its peak for the threshold but it was tiring and time consuming given that we have 12 subimages. So it was grateful to know that Scilab has all its way of making our image processing easier :). As I explored the Image Processing Design (IPD) module of Scilab, I came across the CalculateOtsuThreshold() and SegmentByThreshold() functions. (IPD is available on ATOMS Module Manager of Scilab. You would only need to install it manually.) The first function is able to solve the threshold of the images automatically while the latter is used to binarized the image with the set threshold value
We favor the values higher than the threshold to clearly represent the cells, just don't make it so high that everything would appear black. So in binarizing the image, we add 20 to the calculated threshold value. This resulted to more distinguishable ROI and background. The comparison of using different threshold values can be seen below.
 |
| Converting the image to its binary image with threshold equal to the image threshold value (left) and threshold+20 (right). |
Seen below are all the binarized subimages
However, this image cannot still be used for measurement analysis because this is still 'dirty'. With the use of morphological operation done in the previous activity, the image can be further cleaned up. Understanding the closing, opening and tophat operator, the image can be cleaned nicely. I really had a hard time doing this part because there were a lot of factors to consider. First, was the structuring element to be used then you would then think of the right combination. I understand well the use of the closing and opening operator because its name was already self-explanatory, while the tophat operator is just confusing. So I decided not to use it in this activity :P
 |
| A sample of the cleaned image applying closing operator first (left) then open operator (right). |
I realized that the structuring element that I need to use was a circle, the same shape as our cells. The only problem is the size. I first use the CloseImage() function so that our ROI will fully cover our cells then OpenImage() function was used to separate adjacent cells. This separation was only effective for cells nearly touching each other but for overlapping cells, it just doesn't do much. However, this operator was still useful because this make the excess dirt pixels in the background to be wiped out. See image above :)
Shown below are all the cleaned subimages.
Now, we can now analyze our blobs. SearchBlobs(), another function of IPD, was used to assign numbers to each blob found in the image. Now, each blob can be called separately, thus will able to analyze its features like area.To ensure that the blobs have their own individuality, the ShowImage() function was used resulting to the graph of blobs respresented with different color. And just for the heck of trying, I tried the the bounding box feature, which can all be found in the blob analysis in the IPD module.
The area of each blob can be easily computed using the size or the length function however, the area of each cell cannot be obtained because not all the blobs represent only one cell as what is observed on the blobs on the right portion of the images above. So I establish a code that if it consider the the blob contains two cell, the area will be divided by two, and so on. I completely understand that this procedure will make the estimate inaccurate but this was the best solution that I could think of.
Table below shows the area computed for each blob of each subimages.
From all these data the computed mean and standard deviation were 525.69605 and 94.437235, respectively. The calculated best estimate of the area of the 'cells' is
Isolating the cancer cells from the healthy ones
Using the best estimate, the cancer cells, represent by abnormally large size cells, can be isolated from the normal cells. The image containing both cancer and normal cells is shown below.
Doing the same process in first image, we search for the blobs in this second image. I just processed the image as it is, without any cropping. The binarized image and the cleaned image are shown below
I isolated the cancer cell simply by using the FilterBySize() function of the Scilab with bounds of [431, 620] repesenting the range of the best estimate of the area. The result is as follows.
This looks pretty much right. The cancer cells were removed however other normal cells were also removed. The cells that was taken out was displayed by setting the parameters of FilterBySize() greater than the best estimate bounds.
So, yes! There were normal cell that be removed. The same as the problem previously, there were overlapped cells and since they were big, the code consider it as cancer cell.
I give myself a grade of 10/10. I wasn't able to produce the desired output but I give myself a consolation point for exploring other stuff and the product of real hardwork :)
__________
References
[1] Soriano,
M. "Application of Binary Operations 1 : Blob Analysis." AP 186 Laboratory Manual.
National Institute of Physics, University of the Philippines, Diliman.
2013.
This been an exhaustingly dull weekend and weekdays. The rain just really set the mood a little off. I've been confined inside my room for like countless days, sitting in front of my computer doing my blogs and papers, or listening to music, or watching TV, and just going outside in need for food. And whenever I see bad news concerning the storm, I can't help but feel down.. especially if the news is about your province. Bataan is now under state of calamity and all I can is pray for the safety of my friends and loved ones. Stay safe everyone!